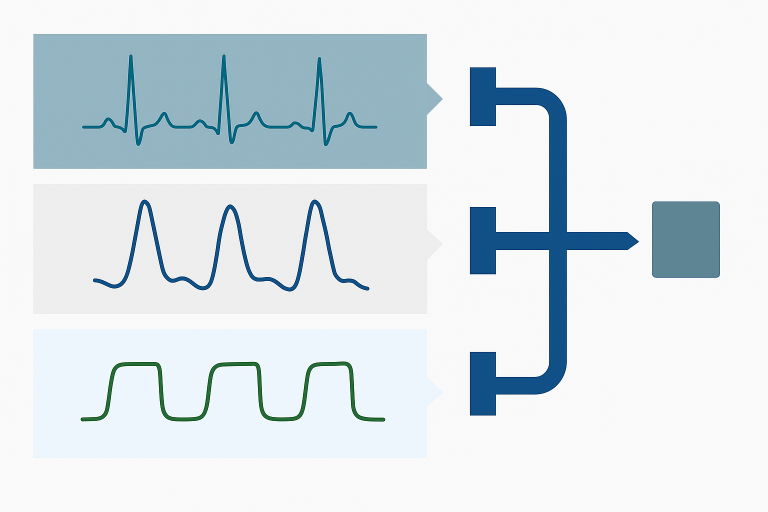
Neural networks offer powerful capabilities for interpreting physiological signals and uncovering subtle patterns that reflect the body’s condition. When properly designed, they can extract meaningful insights that are difficult to capture with traditional methods. However, training neural networks typically requires large volumes of high-quality data—a major challenge in many health and fitness applications, where data collection can be costly, time-consuming, or impractical. To address this, PhysioSense has developed a suite of foundational AI models for various physiological sensors (eg PPG, ECG, etc.) that significantly reduce data requirements while preserving the advantages of deep learning. These models provide a robust starting point for your AI initiatives, accelerating development and enhancing interpretability from limited physiological data.
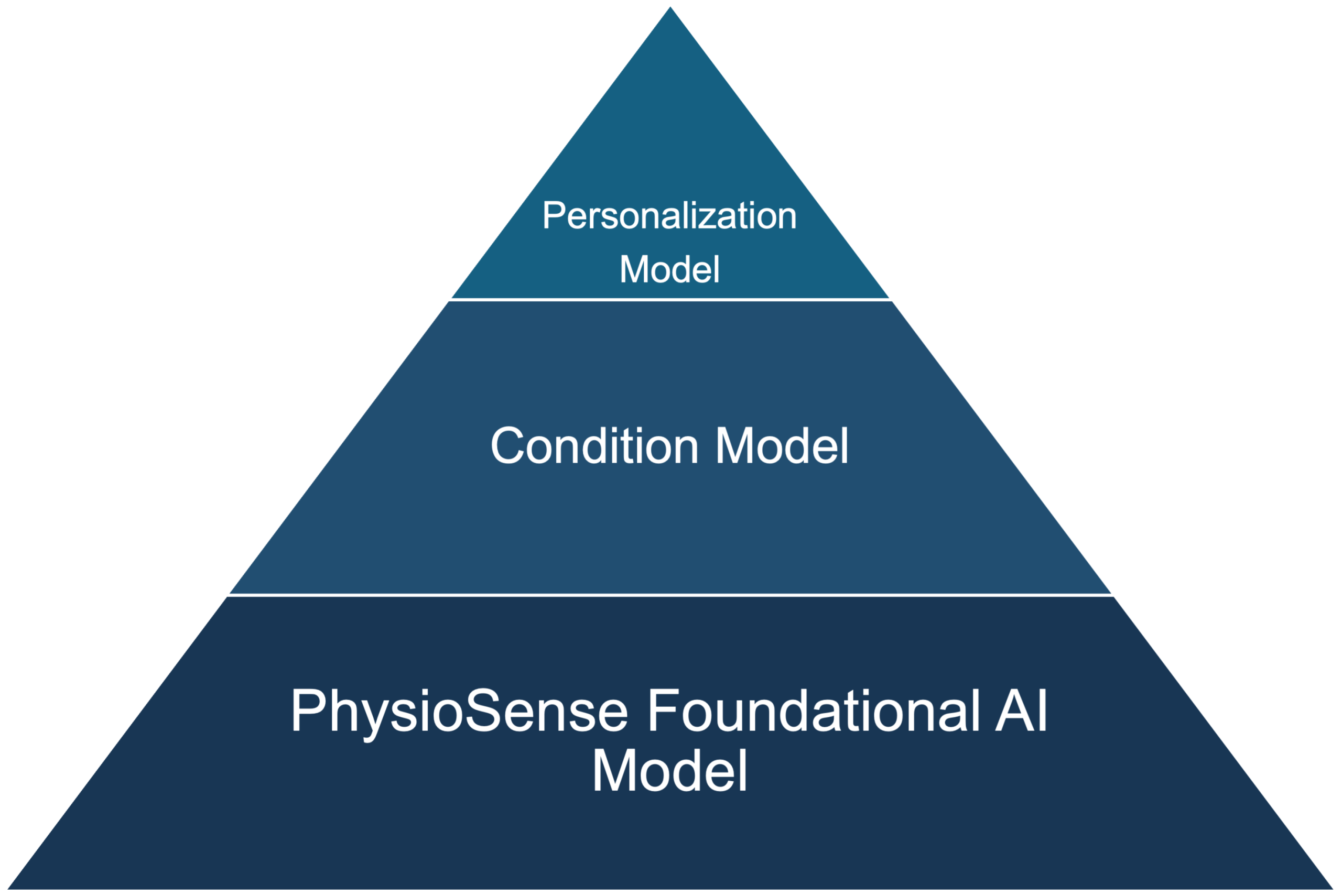
The Foundational Models have been trained on large volumes of real patient data, enabling them to learn the diverse range of valid waveform shapes and signal variations across individuals and conditions. By capturing these low-level signal characteristics, the foundational layer reduces the need for extensive labeled datasets when training models for specific applications.
Building on this base, developers can train Condition Models that map waveform patterns to physiological or clinical states—such as dehydration, sepsis, or diabetes—using significantly less labeled data than would otherwise be required. Finally, Personalization Models can be layered on top to adapt predictions to individual users through calibration or personal history, enabling accurate, real-time interpretation for diverse populations and use cases. This modular, layered approach supports faster development, improved performance, and broader applicability of physiological AI models.
The Foundational Models are accessible via our SensorFusion REST API and PhysioStack SDK.
Subtitle for This Block